Is This the ELISA 2.0 We've Been Waiting For?
The field of Antibody-Oligonucleotide Conjugate (AOC) techniques continues developing at an impressive pace. Following on from our earlier summary of AOC techniques in Scientific Techniques Using Antibody-Oligonucleotide Conjugates, I wanted to highlight a new paper from Nomic Bio with an elegant ELISA update.
The Problem
Traditionally trying to multiplex ELISAs resulted in reagent-driven cross-reactivity—when multiple antibody pairs are mixed, non-specific binding events increase exponentially. This has limited most multiplexed sandwich assays to ~10-50 targets, despite the desperate need for broader protein panels in drug discovery and diagnostics.
The Solution
The nELISA platform from Nomic Bio uses a clever DNA-mediated approach called CLAMP (Colocalized-by-Linkage Assays on Microparticles). Detection antibodies are pre-assembled onto their corresponding capture antibody-coated beads using flexible ssDNA tethers. No antibody is in solution and the spatial confinement prevents noncognate interactions. Signal is generated via a complex process using fluorescent DNA oligos. The assay is run in 384-well plates with readout via flow cytometry.
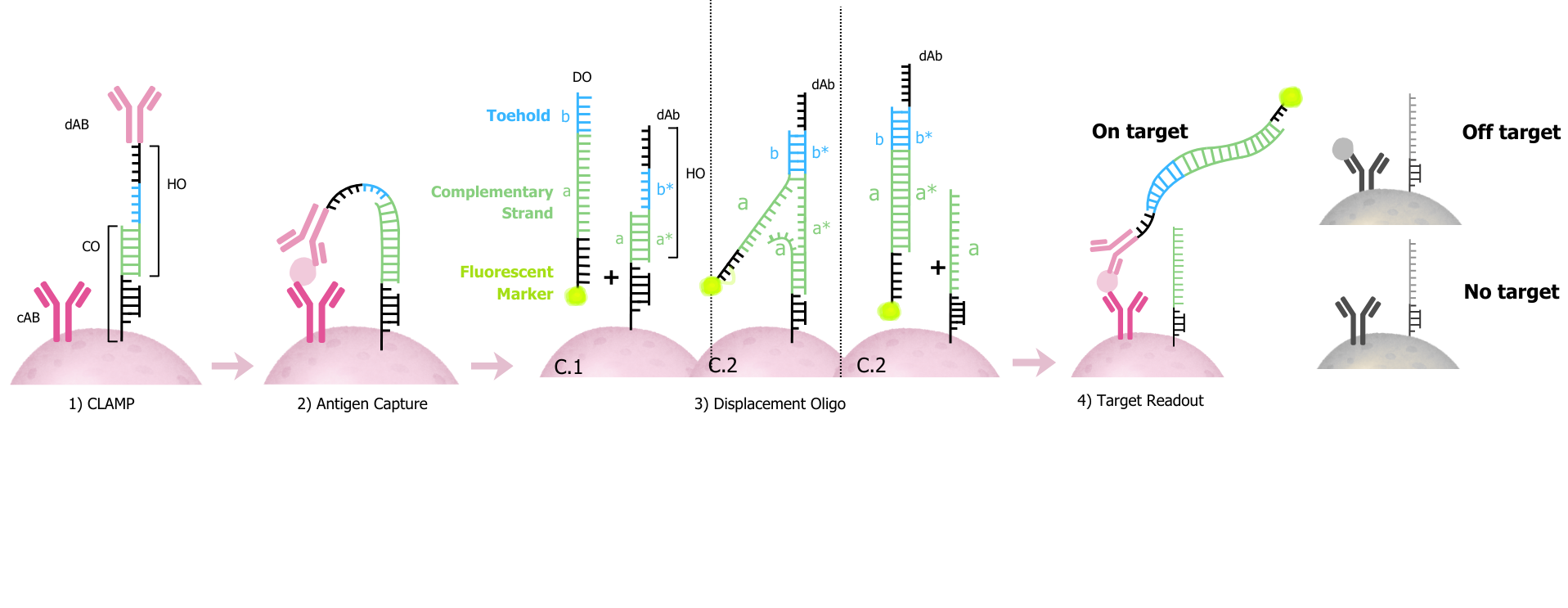
The CLAMP workflow: From antibody setup through displacement oligo to target readout
Each barcoded bead functions like a pre-assembled ELISA, specifically targeting the protein of interest:
1. CLAMP Setup
The detection antibody (dAb) is coupled to a short "hook" oligo (HO) which is held at the bead surface by hybridizing to an immobilized capture oligo (CO). Note: cAb signifies the capture antibody.
2. Antigen Capture
The beads are mixed with the biological sample forming a sandwich complex only when the protein is present, bridging the capture side and the dAb.
3. Displacement Step
After washing, a fluorescent displacement oligo (DO) is introduced. DO uses a toehold to kick HO off the CO through strand displacement: the DO toehold segment (b) binds the exposed complementary region (b*) on HO, which then drives the exchange—CO's segment (a) is "unzipped" from HO's (a*), and DO's (a) takes its place. The result is that HO is released from CO and becomes fluorescently tagged via the DO.
4. Readout
If the target antigen is present, the sandwich complexes stay anchored on the surface and retain the fluorescent label. If the target is absent (or binding is non-specific), the dAb–DO species is removed during washing, leaving little to no signal.
Key Achievements
- 191-plex inflammation panel with sub-picogram/mL sensitivity across 7 orders of magnitude (calibration standards with recombinant antigens for all of them)
- Profiled 7,392 PBMC samples generating ~1.4 million protein measurements in under a week
- Seamlessly integrated with Cell Painting for multimodal phenotypic screening
Kudos to Milad Dagher and the team at Nomic Bio, alongside David Juncker lab at McGill University and collaborators at the Broad Institute including Anne Carpenter's imaging platform.
What Would It Take to Truly Replace ELISA?
The technology is impressive, but widespread adoption will require:
- Flexibility for researchers to assemble and customize their own panels and go from singleplex to multiplex easily
- Compatibility with equipment already in most labs
- Broad availability of AOC reagents
The more accessible these conjugates become, the faster this kind of technology can move from specialist platforms to the everyday bench. AbOliGo's revolutionary approach for flexible selection alongside rapid production of antibody-oligonucleotide conjugates (AOCs) at microgram scale is perfect to help accelerate adoption of these platforms.