Can DIY Microscopy Finally Democratize Spatial Biology?
The field of Antibody-Oligonucleotide Conjugate (AOC) techniques continues developing at an impressive pace! Following up our earlier summary of AOC techniques in Scientific Techniques Using Antibody-Oligonucleotide Conjugates, I wanted to highlight a paper that tackles one of the biggest barriers to adoption: cost, accessibility and flexibility.
The Problem
Multiplexed imaging platforms have transformed tissue biology, but commercial systems remain prohibitively expensive and locked to proprietary reagents. This limits access to well-funded core facilities and restricts researchers who need to customize protocols or integrate novel reagents.
The Solution
Fluid-Squid (Simplifying QUantitative Imaging platform Development) combines the open-source Squid microscopy platform with automated fluidics for cyclic imaging, all for around $30k in materials. The compact system has been field-tested from Stanford to Liberia, proving its robustness in diverse settings.
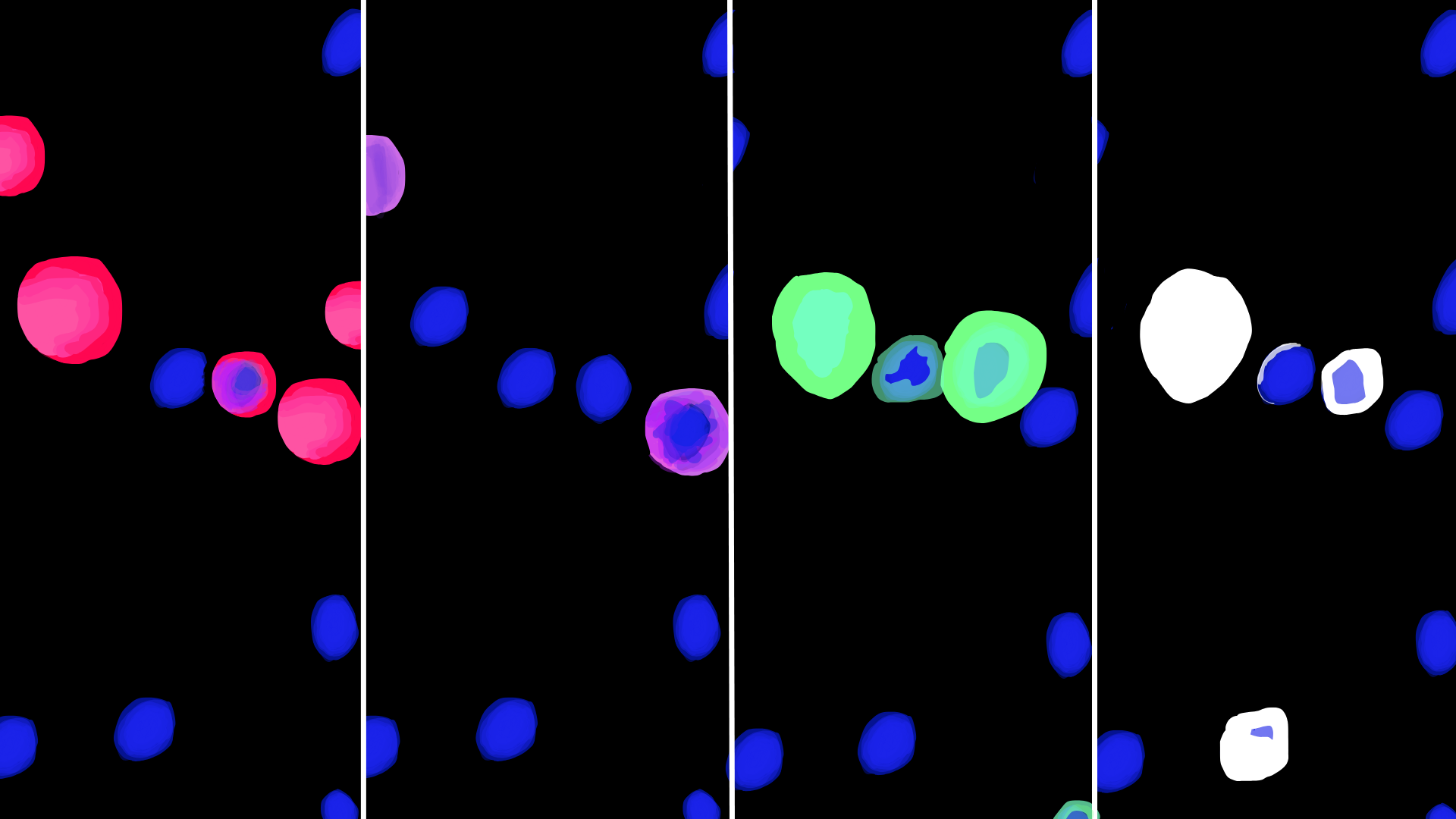
Multiplexed AOC imaging showing distinct cell populations visualized with different fluorescent markers
Key Achievements
- 36-plex AOC imaging of human intestine tissue across a 4.5mm × 4.5mm area at single-cell resolution
- Extended workflows to dissociated single cells (PBMCs and splenocytes) with a 39 AOC panel
- Demonstrated lyophilization of AOC panels, critical for field deployment where cold-chain storage isn't feasible
- Developed AOC conjugation strategy enabling simultaneous staining and rapid titration of 40+ marker panels in single experiments
What Will It Take for Widespread Adoption?
The hardware democratization is impressive, but the real bottleneck may shift to reagents. Labs building custom panels still need access to AOCs and the broader availability of ready-to-use AOC reagents could dramatically lower the barrier for prototyping spatial proteomics experiments without being restricted to a specific protocol.